Sophia Stone
 |
ASSOCIATE PROFESSOR BSc (York University, 1998), PhD (York University, 2003) NSERC Postdoctoral Fellow (University of California-Davis, 2004) International Human Frontier Science Program Long-term Fellow (UC-Davis, 2006) |
|
| Teaching & Research plant developmental biology, molecular and cell biology, Arabidopsis thaliana, protein ubiquitination, regulated proteolysis, kinase signaling
esearch in my laboratory focuses on understanding the regulatory role of protein ubiquitination in plant development. The ubiquitination pathway covalently attaches the small ubiquitin molecule to protein substrates through the sequential action of three enzymes, the E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugatin The goal of my research is to define the function of this important pathway in plant development using the small crucifer Arabidopsis thaliana as a model system. I am particularly interested in the function of the RING class of E3 ligases. We utilize a number of molecular, genetic, biochemical and proteomic approaches to determine the role of RING E3 ligases, to identify targets and to determine the consequence of ubiquitination in relation to plant development. Of the over 1000 E3s found in the Arabidopsis proteome, 469 are RING E3 ligases.
The RING family represents eight different types of E3 ligases based on variations within the E2 binding RING domain, or thirty different subgroups based on the presence of other types of motifs/domains. Efforts to characterize selected novel and plant specific sub-groups of GRADUATE STUDENTS and POST-DOCs |
||
Selected Publications Chen YT, Liu H, Stone S, Callis J. (2013) ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. Jun 6. doi: 10.1111/tpj.12259. [Epub ahead of print] Liu H, Stone SL (2013) Cytoplasmic Degradation of the Arabidopsis Transcription Factor ABSCISIC ACID INSENSITIVE 5 is Mediated by the RING-type E3 Ligase KEEP ON GOING J. Biol. Chem. published May 29, 2013 as doi:10.1074/jbc.M113.46536 Lyzenga WJ, Liu H, Schofield A, Muise-Hennessey A, Stone SL. (2013) Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J Exp Bot. 2013 May 8. [Epub ahead of print] Lyzenga WJ, Booth JK, Stone SL. (2012) The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 71(1):23-34. Liu H, Stone SL. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22(8):2630-41. Prasad ME, Schofield A, Lyzenga W, Liu H, Stone SL. (2010) Arabidopsis RING E3 ligase XBAT32 regulates lateral root production through its role in ethylene biosynthesis. Plant Physiol. 153(4):1587-96. Stone SL, Williams LA, Farmer L, Vierstra RD and Callis J (2006) KEEP ON GOING, a RING E3 Ligase Essential for Arabidopsis Growth and Development is involved in Abscisic Acid Signaling. Plant Cell 18(12):3415-28 Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau O-S, Deng XW and Callis J (2005) Genome Analysis and Functional Characterization of the E2 and RING domain E3 ligase Ubiquitination Enzymes of Arabidopsis thaliana. Plant Physiol. 139: 1597-1611. Links |


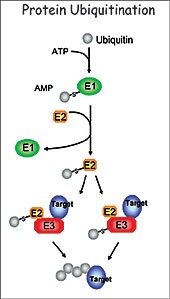 g enzyme), and E3 (ubiquitin ligase). Specificity of the pathway is mainly governed by a large number of diverse E3 ligases that recognize the appropriate substrates. The best-characterized consequence of ubiquitination is degradation of the targeted protein by the 26S proteasome. This highly conserved pathway removes abnormal proteins and controls the abundance of important regulatory proteins, thus allowing for the control of numerous cellular networks and regulation of a wide range of developmental processes.
g enzyme), and E3 (ubiquitin ligase). Specificity of the pathway is mainly governed by a large number of diverse E3 ligases that recognize the appropriate substrates. The best-characterized consequence of ubiquitination is degradation of the targeted protein by the 26S proteasome. This highly conserved pathway removes abnormal proteins and controls the abundance of important regulatory proteins, thus allowing for the control of numerous cellular networks and regulation of a wide range of developmental processes. RING E3s include demonstrating ligase activity, isolating protein substrates, determining expression patterns and analysis of Arabidopsis mutants to determine function.
RING E3s include demonstrating ligase activity, isolating protein substrates, determining expression patterns and analysis of Arabidopsis mutants to determine function.